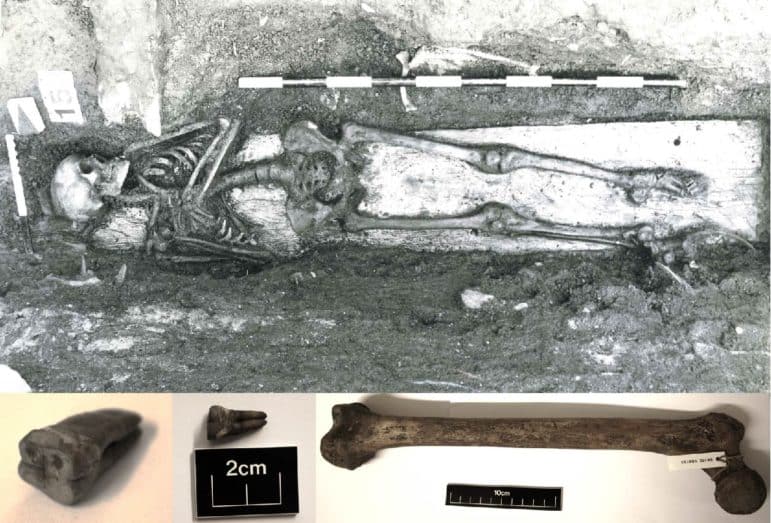
Researchers have detected Salmonella Paratyphi C, which causes enteric fever, in a 800-year-old human skeleton from Trondheim, Norway. This suggests the potentially lethal disease, more common in hot countries, was present in medieval Europe.
Now scientists are speculating that the evolution of enteric fever could be linked to the domestication of pigs across northern Europe.
“This is the first time that any Salmonella have been found in old human remains in Europe…”
Mark Achtman, professor at Warwick University’s Medical School, and his team analyzed bacterial DNA found in the teeth and bones of the skeleton of a young woman who is believed to have migrated to Trondheim from the northernmost areas of Scandinavia or Northwest Russia by her early teens only to die there around the age of 19 to 24.
They reconstructed a genome of Salmonella Paratyphi C, which causes enteric fever in areas of poor sanitation and lack of clean drinking water. Their discovery indicates that the young Norwegian died of this disease and suggests that these bacteria have long caused enteric fever across northern Europe.
“Paratyphi C is very rare today in Europe and North America except for occasional travelers from South and East Asia or Africa, where the disease is more common,” says Achtman.
“This is the first time that any Salmonella have been found in old human remains in Europe, which is surprising because other Salmonella are more common today, including Salmonella causing typhoid fever, called Typhi, and Salmonella causing food poisoning.”
The new results include comparative analyses of the Paratyphi C genome from the skeleton against modern Salmonella genome sequences from the online database EnteroBase. This revealed that Paratyphi C represents the evolutionary descendants of a common ancestor, or clade, within the Para C lineage. The Para C Lineage includes Choleraesuis, which causes septicemia in pigs and boar, and Typhisuis, which causes epidemic swine salmonellosis (chronic paratyphoid) in domestic pigs.
These different host specificities likely evolved in Europe over the last 4,000 years and coincide with the timing of pig domestication in Europe.
Our bodies can make Salmonella more toxic
According to historical records, bacterial infections have long afflicted humans, yet genomic analyses of living bacterial pathogens routinely estimate a date for the most recent common ancestor of no more than a few centuries. In general, evolutionary trees contain a stem group, which may include lineages that are now rare or extinct, as well as the crown group of living organisms. Historical reconstructions based only on the crown group ignore the older sub-lineages in the stem group and thereby provide an incomplete picture of the older evolutionary history of the pathogen.
In contrast, analyses of ancient DNA such as the Paratyphi C genome can shed light on additional millennia of bacterial pathogen evolution that occurred prior to the origin of the crown group.
Repeat food poisoning may lead to chronic intestine trouble
Achtman adds: “Using EnteroBase we were able to define the Para C lineage from 50,000 modern Salmonella enterica genomes and find that over its 3,000-year history only a few genomic changes occurred within the Para C lineage.
“As well as reshaping our understanding of Salmonella enterica, our research has triggered intriguing speculations about historical host jumps during the Neolithic period between humans and their domesticated animals.”
The findings appear in Current Biology.
Source: University of Warwick