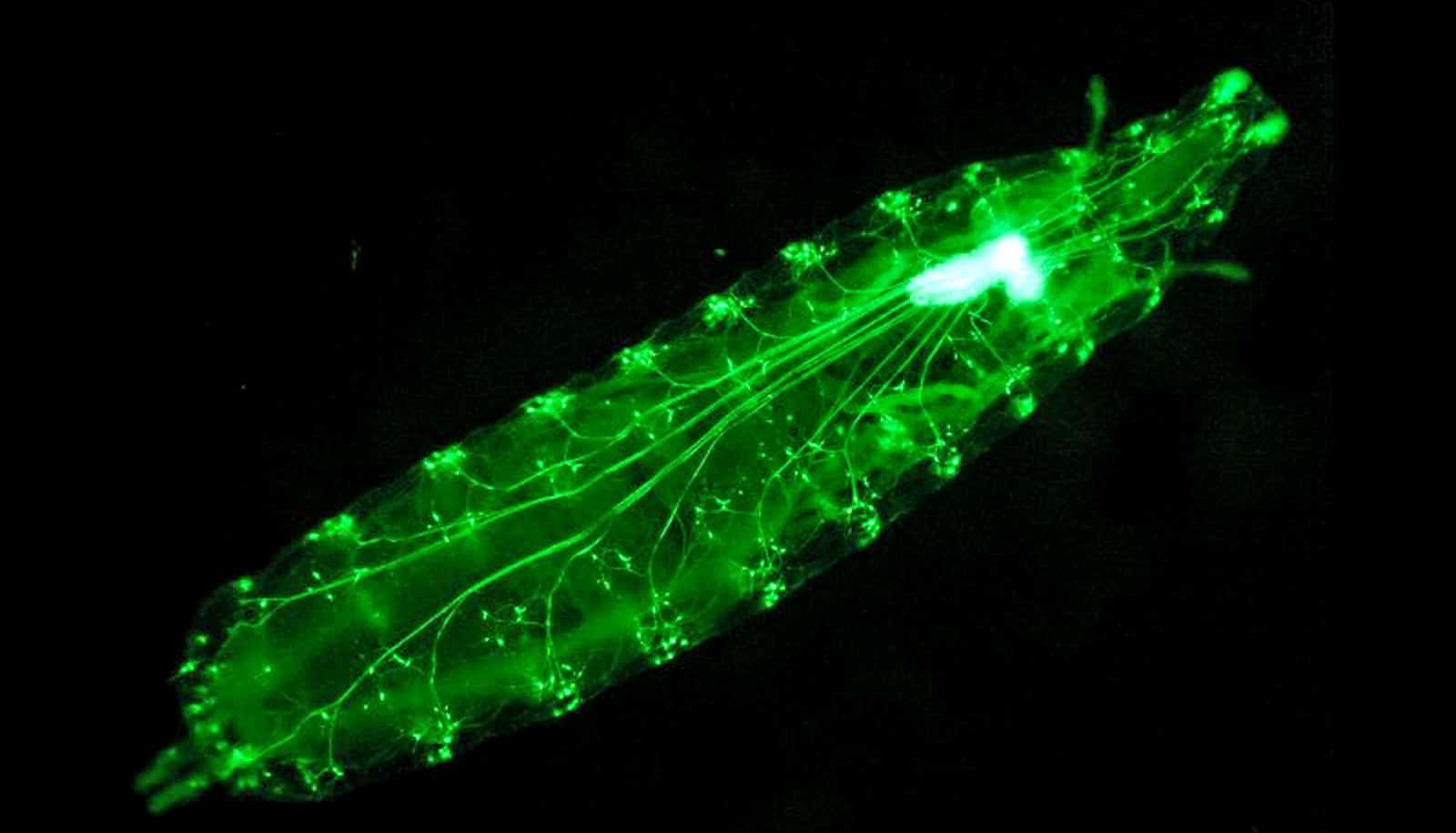
A tool that finds and labels neurons during development has led to the discovery of new cell types in the visual system of flies, according to a new study.
The study, published in the Proceedings of the National Academy of Sciences, combines single-cell sequencing data with a new algorithm to identify pairs of genes that point to previously unknown cells in the brains of fruit flies.
Researchers have long used fruit flies (also known as Drosophila) as a model organism to study fundamental questions about the development and function of the brain. Instead of the 86 billion neurons found in humans, fruit flies have about 100,000 neurons—making research into the brain a more manageable, yet still complex, endeavor.
The use of genetic tools that can distinguish different types of cells in fruit flies has revolutionized the study of neural circuits in the brain, allowing scientists to understand circuit development, function, and behavior in a precise manner.
“A hallmark of the central nervous system is the diversity of different cell types that are responsible for so many different functions,” says senior author Claude Desplan, professor of biology and neural science at New York University.
Previous research in Desplan’s lab used single-cell sequencing to determine that there are approximately 200 cell types in the developing fly’s visual system. Single-cell sequencing reveals gene expression, so when cells have the same gene expression patterns, they are likely doing the same job and are therefore the same cell type.
Scientists could identify roughly half of the 200 cell types in the developing fly’s visual system based on their gene expression and prior studies, but they lacked a way to more easily study and label the other 100 cell types.
Existing tools that allowed precise manipulation of neural circuits of adult fruit flies often failed to label the same neurons during development, rendering these tools unfit to study cells in the developing brain.
“Moreover, the previous approach to identifying cell types involves laborious testing of numerous gene candidate combinations. We knew we needed a much more efficient approach to label specific cell types, and were able to tap into the growing amount of single-cell sequencing data that is available,” says first author Yu-Chieh David Chen, a postdoctoral associate in the biology department.
Chen and his colleagues created a tool that takes advantage of the extensive single-cell sequencing data for the developing fly visual system to identify genes—and combinations of genes—that are exclusively expressed in certain cell types.
To find a cell type, researchers typically look for genetic markers, or single genes that are specific to a cell type. But often a gene will be expressed in multiple cell types, making it difficult to use one gene to differentiate between them. The new tool uses a slightly different approach: finding two genes that overlap only in one cell type.
By feeding single-cell RNA sequencing data into an algorithm they created, the researchers systematically identified pairs of genes that are uniquely expressed in the majority of cell types in the fruit fly’s visual system at multiple stages of development. One such gene pair led to the discovery of MeSps, a brand-new cell type.
“Despite a long history of studying the fruit fly’s visual system, we had never seen this cell type before,” says Chen.
While future research will delve into the development and function of MeSps—for instance, whether it detects color, motion, or other features of light—this avenue of research will be made possible by the new tools.
The researchers note that their tools can also be used to study other systems beyond vision in the developing fly, as long as single-cell data are available. Moreover, their logic of finding marker gene pairs instead of one single marker gene can be applied in research in other species.
“Instead of looking for a single good marker gene, a simple tweak of just looking at two genes can achieve high cell-type specificity,” says Chen.
“This pioneering and efficient approach provides exceptional tools for the field of neuroscience to investigate developmental questions with high precision,” says Desplan.
Additional coauthors are from the University of Missouri-Kansas City, the University of Toronto-Mississauga, and NYU.
The National Institutes of Health supported the work.
Source: NYU