Researchers have created protein origami, or nanostructures, in the shapes of triangles and squares using stable protein building blocks.
The work draws inspiration from DNA origami, in which folding DNA forms nanostructures.
The protein nanostructures can endure high temperatures and harsh chemical conditions, neither of which is possible with DNA-based nanostructures. In the future, these protein nanostructures could help improve sensing capabilities, speeding chemical reactions, drug delivery, and other applications.
When trying to create protein nanostructures suited for particular applications, researchers typically make modifications to existing protein structures, such as virus particles. However, the shapes of nanostructures that they can make using this approach are limited to what nature provides.
Now, researchers have developed a bottom-up approach to build 2D nanostructures, essentially starting from scratch.
“Building something that nature has not offered is more exciting,” says Fuzhong Zhang, associate professor of energy, environmental, and chemical engineering at Washington University in St. Louis. “We took individually folded proteins and used them as building blocks, then assembled them together piece by piece so that we can create tailored nanostructures.”
Building blocks form protein origami
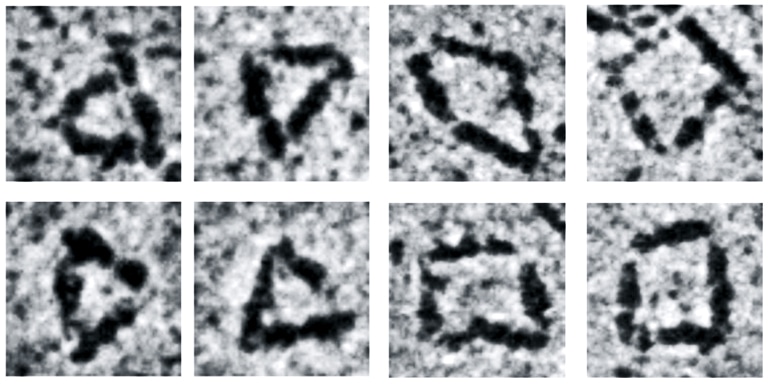
Using synthetic biology approaches, Zhang’s team first biosynthesized rod-shaped protein building blocks, similar in shape to a pencil but only 12 nanometers long.
Then, they connected these building blocks together through reactive protein domains genetically fused to the ends of each of the rods, forming triangles with three rods and squares with four rods. These reactive protein domains are known as split inteins, which are not new to Zhang’s lab—they are the same tools that his group uses to make high-strength synthetic spider silk and synthetic replicas of the adhesive mussel foot proteins.
In both cases, these split intein groups enable the production of large proteins that make the synthetic spider silk tougher and stronger and the mussel foot proteins stickier. In this case, they enable the construction of novel nanostructures.
The researchers worked with Rohit Pappu, professor of engineering, professor of biomedical engineering, and an expert in the biophysics of intrinsically disordered proteins, phase transitions, and protein folding, to “understand how the protein sequence at the connections determines the flexibility of these nanostructures and helped us to predict protein sequences to better control the flexibility and geometry of nanostructures,” Zhang says.
The collaboration simplified a very complex process.
“Once we understood the design strategy, the work is fairly straightforward and quite fun to do,” Zhang says. “We just controlled the different functional groups, then they controlled the shapes.”
Tough, tiny structures
Due to the versatile functionality of proteins, these nanostructures potentially could be useful as scaffolds to assemble various nanomaterials. To test this idea, the team assembled 1-nanometer gold nanoparticles precisely at the vertices of the triangle. Using a state-of-the-art electron microscope, researchers could see both the protein triangles and the gold nanoparticles assembled to the vertices of the triangles.
To test the stability of these protein nanostructures, the team exposed them to high temperatures, up to 98 degrees Celsius, to chemicals such as guanidium hydrochloride, and to organic solvents such as acetone. While these conditions generally destroy protein structures, the structures from Zhang’s lab stayed intact. This ultra-stability could enable more nanoscale applications that are difficult or not possible using nanostructures made from DNA or other proteins, Zhang says.
Next, the team is working to use these protein nanostructures to develop improved plasmonic sensors.
“Exploiting the interplay between highly stable structural building blocks and intrinsically disordered or flexible regions provides a novel route to designing nanostructures with customizable features for a variety of applications in synthetic biology and biomedical sciences,” says Pappu.
The results appear in Nature Communications. The Office of Naval Research, NASA Space Technology Research Fellowship, Human Frontier Science Program, and the National Science Foundation provided funding for the work.