A high-efficiency gene-editing tool can get fungi to produce significantly more natural compounds, including some previously unknown to the scientific community, say researchers.
Using the approach that simultaneously modifies multiple sites in fungal genomes, Rice University chemical and biomolecular engineer Xue Sherry Gao and collaborators coax fungi into revealing their best-kept secrets, ramping up the pace of new drug discovery.
It is the first time that the technique, multiplex base-editing (MBE), has been deployed as a tool for mining fungal genomes for medically useful compounds. Compared to single-gene editing, the MBE platform reduces the research timeline by over 80% in equivalent experimental settings, from an estimated three months to roughly two weeks.
Fungi and other organisms produce bioactive small molecules such as penicillin to protect themselves from disease agents. These bioactive natural products (NPs) can be used as drugs or as molecular blueprints for designing new drugs.
The study appears in the Journal of the American Chemical Society.
Gene-editing fungi
Base-editing refers to the use of CRISPR-based tools in order to modify a rung in the spiral ladder of DNA known as a base pair. Previously, gene modifications using base-editing had to be carried out one at a time, making the research process more time-consuming. “We created a new machinery that enables base-editing to work on multiple genomic sites, hence the ‘multiplex,'” Gao says.
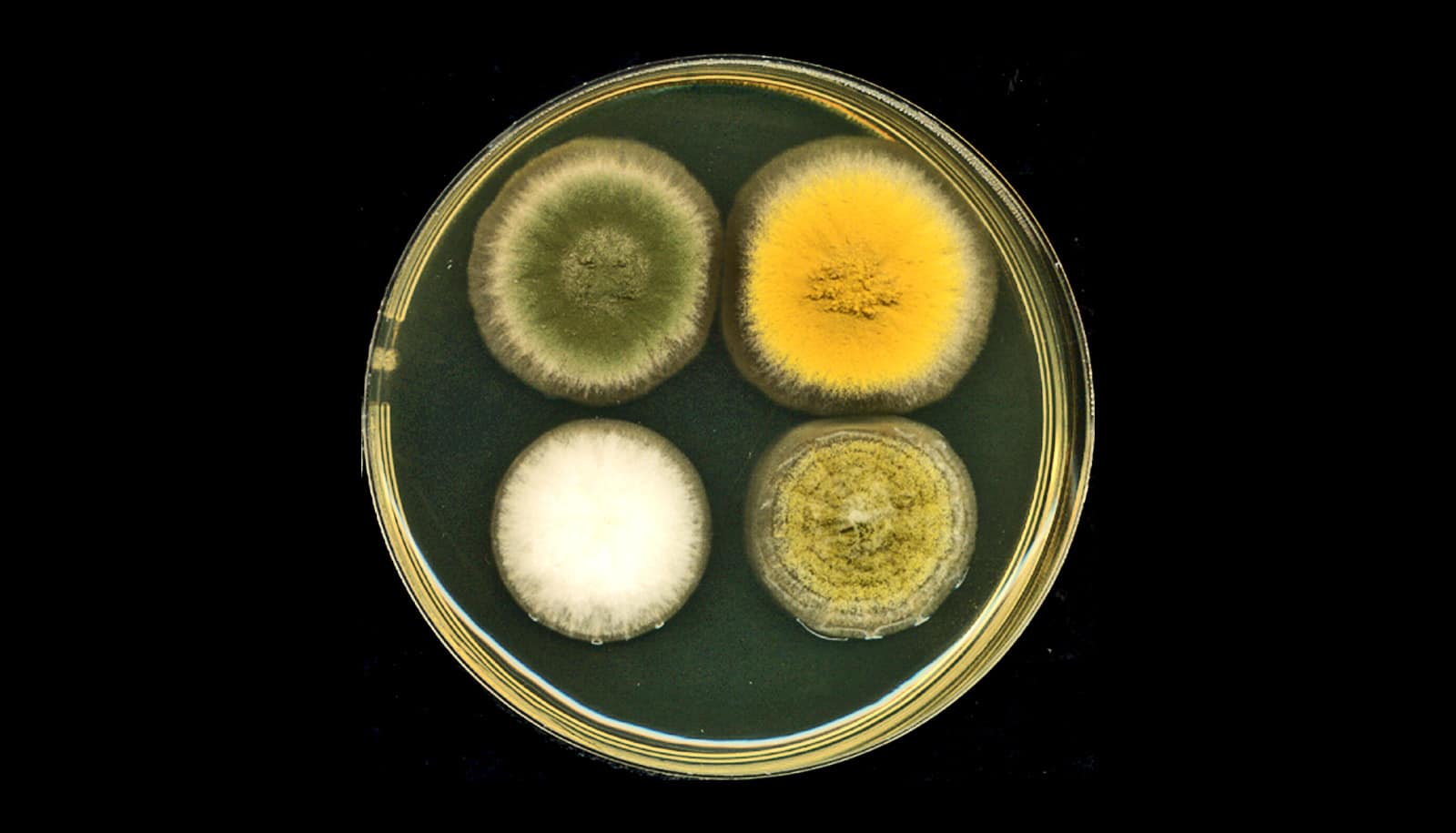
Gao and her team first tested the efficacy of their new base-editing platform by targeting genes encoding for pigment in a fungal strain known as Aspergillus nidulans. The effectiveness and precision of MBE-enabled genome edits was readily visible in the changed color displayed by A. nidulans colonies.
‘Cryptic’ genes
“To me, the fungal genome is a treasure,” Gao says, referring to the significant medical potential of fungi-derived compounds. “However, under most circumstances, fungi ‘keep to themselves’ in the laboratory and don’t produce the bioactive small molecules we are looking for. In other words, the majority of genes or biosynthetic gene clusters of interest to us are ‘cryptic,’ meaning they do not express their full biosynthetic potential.
“The genetic, epigenetic, and environmental factors that instruct organisms to produce these medically useful compounds are extremely complicated in fungi,” Gao says. Enabled by the MBE platform, her team can easily delete several of the regulatory genes that restrict the production of bioactive small molecules. “We can observe the synergistic effects of eliminating those factors that make the biosynthetic machinery silent,” she says.
Disinhibited, the engineered fungal strains produce more bioactive molecules, each with their own distinct chemical profiles. Five of the 30 NPs generated in one assay were new, never-before-reported compounds.
“These compounds could be useful antibiotics or anticancer drugs,” Gao says. “We are in the process of figuring out what the biological functions of these compounds are and we are collaborating with groups in the Baylor College of Medicine on pharmacological small-molecule drug discovery.”
Gao’s research plumbs fungal genomes in search of gene clusters that synthesize NPs. “Approximately 50% of clinical drugs approved by the US Food and Drug Administration are NPs or NP-derivatives,” and fungi-derived NPs “are an essential pharmaceutical source,” she says. Penicillin, lovastatin, and cyclosporine are some examples of drugs derived from fungal NPs.
The National Institutes of Health and the Robert A. Welch Foundation supported the research.
Source: Rice University