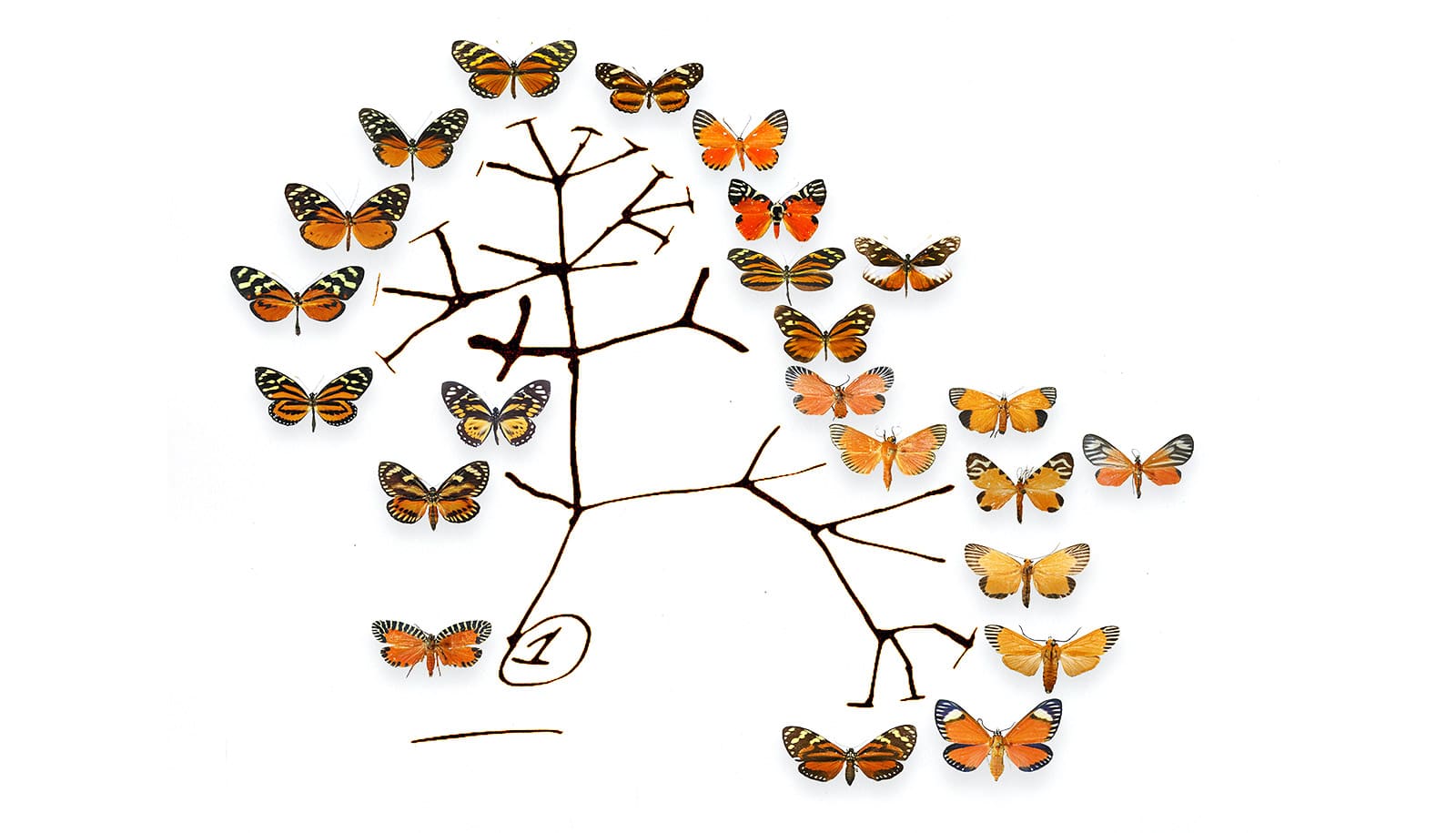
Researchers say they’ve solved the question of what genetic information can help explain past extinctions and the evolution of new and distinct species.
In a new paper in the journal Nature, they argue that long-used approaches for reconstructing evolutionary paths are deeply flawed.
While paleontology provides insights on how and why patterns of biodiversity have changed over geological time, fossils of many organisms are too scant to say anything, says Stilianos Louca, an assistant professor in the biology department and member of the University of Oregon’s Institute of Ecology and Evolution. An alternative approach that relies on signals of identifiable changes in an organism’s genetic makeup also can be misleading.
“…I thought I had a really good sense of how these models worked. I was wrong.”
“Our finding casts serious doubts over literally thousands of studies that use phylogenetic trees of extant data to reconstruct the diversification history of taxa, especially for those taxa where fossils are rare, or that found correlations between environmental factors such as changing global temperatures and species extinction rates,” Louca says, using a term for populations of one or more organisms that form a single unit.
Louca and Matthew W. Pennell, an evolutionary biologist at the University of British Columbia, offer a way forward with their mathematical model. It introduces alternative variables to identify long-term evolutionary scenarios from phylogenetic data, or data on evolutionary development and diversification.
“I have been working with these traditional types of models for a decade now,” Pennell says. “I am one of the lead developers of a popular software package for estimating diversification rates from phylogenetic trees. And, as such, I thought I had a really good sense of how these models worked. I was wrong.”
Long-used methods have extracted information about evolution from existing data or still-living organisms, using variants of a mathematical birth-death process. These, however, cannot isolate information about both speciation and extinction rates, especially for a majority of taxa, such as bacteria, that have left no fossil record.
The paleontological approach estimates the number of species that have appeared and disappeared in various intervals based on discovered fossils and their estimated minimum and maximum ages. In the phylogenetic approach, information is drawn from evolutionary relationships between existing species, using mostly genetic data, and structured in phylogenetic trees known as “timetrees.” This is often done by finding a speciation-extinction scenario that most likely generates a phylogenetic tree.
“While an impressive suite of computational methods has been developed over the past decades for extracting whatever information is left, until now we lacked a good understanding of exactly what information is left in these trees and what information is forever lost,” Louca says.
Louca and Pennell’s modeling clarifies what variables can be extracted from timetrees under the generalized birth-death model.
“We suggest that measuring and modeling these identifiable variables offers a more robust way to study historical diversification dynamics,” they write in the paper. “Our findings also make clear that paleontological data will continue to be crucial for answering some macroevolutionary questions.”
The results, Louca says, do not invalidate the theory of evolution itself. They do, however, put constraints on what type of information can be extracted from genetic data to reconstruct evolution’s path.
Louca had support from a University of Oregon start-up grant. A grant from Canada’s Natural Sciences and Engineering Research Council funded Pennell’s contributions.
Source: University of Oregon