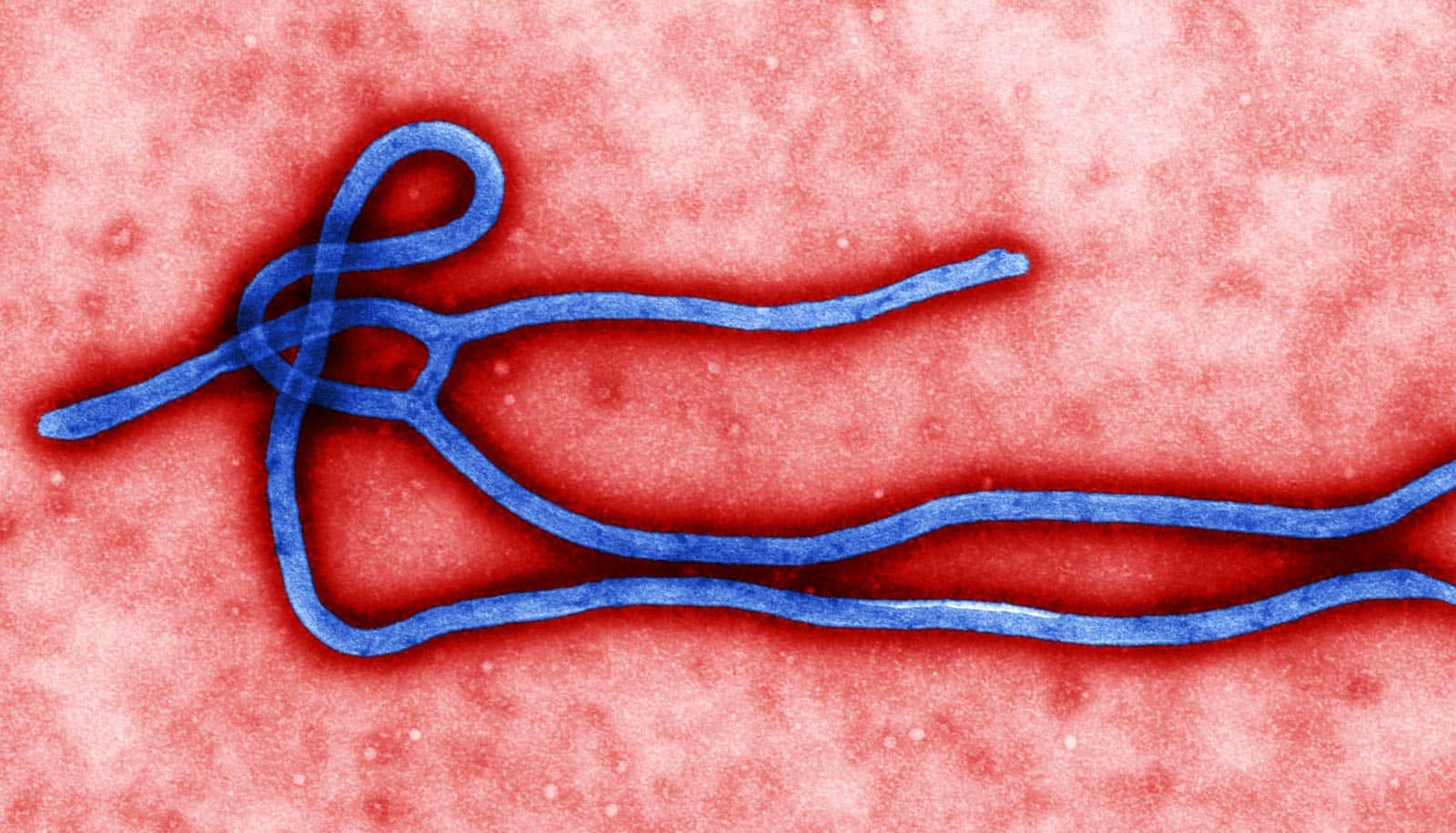
At the start of the epidemic in West Africa, the Ebola virus did not change as rapidly as thought at the time. New research explains why scientists misjudged it.
The culprit is probably methodological biases, according to research led by Tanja Stadler, a professor in ETH Zurich’s department of biosystems science and engineering in Basel. The work appears in PNAS.
When Ebola developed into an epidemic in 2014, an international team of scientists estimated that the pathogen’s genome would change on average every 9.5 days, based on virus samples and computer simulations. This estimate marks an atypically high rate of change. Normally, the Ebola virus genome only mutates at just under half that speed. The high mutation rate led to fears at the time that if the virus rapidly altered, it could also quickly become more virulent.
However, in later studies, researchers evaluating much larger numbers of virus samples could not confirm the high rate. They showed that when viewed over the whole epidemic, the pathogen only changed at its typical slow speed.
The new research shows that the high estimated mutation rates at the start of the epidemic were due to the limited number of virus samples at the time in combination with the computer models scientists used, which calculate the estimates using genetic data from virus samples and from underlying assumptions.
“The smaller the amount of genetic data available for a model, the bigger the influence of the underlying model assumptions on the end result,” explains Stadler.
Ebola RNA lingers in semen longer than expected
Current computer models, however, do not simplify reality as much as those used a few years ago, and they are less heavily influenced by the underlying assumptions, says Stadler. For example, the new models no longer assume that everyone infected has the same probability of passing on the pathogen to other people; instead, they take into account different population structures.
While the new models—the Stadler group is developing some—are more complex and require a lot more computation, they provide more accurate results even at the start of an epidemic, when very little genetic data is available. New calculations by the ETH scientists with the genetic data from 2014 show this increase in accuracy.
Source: ETH Zurich